Chapter 8 - adding new genomes
So we've got a new genome, and we can build a sketch for it. Let's add it into our comparison, so we're building comparison matrix for four genomes, and not just three!
To add this new genome in to the comparison, all you need to do is add
the sketch into the compare_genomes input, and snakemake will
automatically locate the associated genome file and run
sketch_genome on it (as in the previous chapter), and then run
compare_genomes on it. snakemake will take care of the rest!
rule sketch_genome:
input:
"genomes/{accession}.fna.gz",
output:
"{accession}.fna.gz.sig",
shell: """
sourmash sketch dna -p k=31 {input} --name-from-first
"""
rule compare_genomes:
input:
"GCF_000017325.1.fna.gz.sig",
"GCF_000020225.1.fna.gz.sig",
"GCF_000021665.1.fna.gz.sig",
"GCF_008423265.1.fna.gz.sig",
output:
"compare.mat"
shell: """
sourmash compare {input} -o {output}
"""
rule plot_comparison:
message: "compare all input genomes using sourmash"
input:
"compare.mat"
output:
"compare.mat.matrix.png"
shell: """
sourmash plot {input}
"""
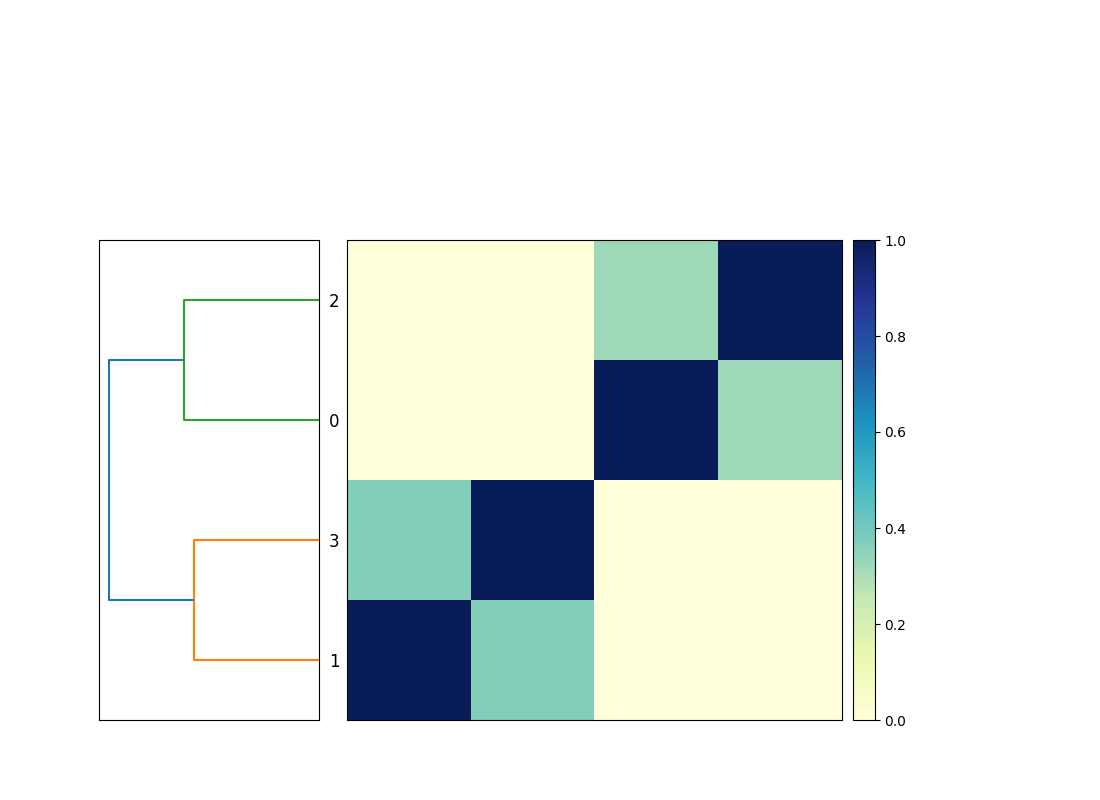
Now when you run snakemake -j 3 plot_comparison you will get a
compare.mat.matrix.png file that contains a 4x4 matrix! (See Figure.)
Note that the workflow diagram has now expanded to include our fourth genome, too!