Khmer Error Trimming
Why (or why not) do k-mer trimming?
Even after quality trimming with Trimmomatic, our reads will still contain errors. Why?
First, Trimmomatic trims based solely on the quality score, which is a statistical statement about the correctness of a base - a Q score of 30 means that, of 1000 bases with that Q score, 1 of those bases will be wrong. So, a base can have a high Q score and still be wrong! (and many bases will have a low Q score and still be correct)
Second, we trimmed very lightly - only bases that had a very low quality were removed. This was intentional because with assembly, you want to retain as much coverage as possible, and the assembler will generally figure out what the “correct” base is from the coverage.
An alternative to trimming based on the quality scores is to trim based on k-mer abundance - this is known as k-mer spectral error trimming. K-mer spectral error trimming always beats quality score trimming in terms of eliminating errors; e.g. look at this table from Zhang et al., 2014:
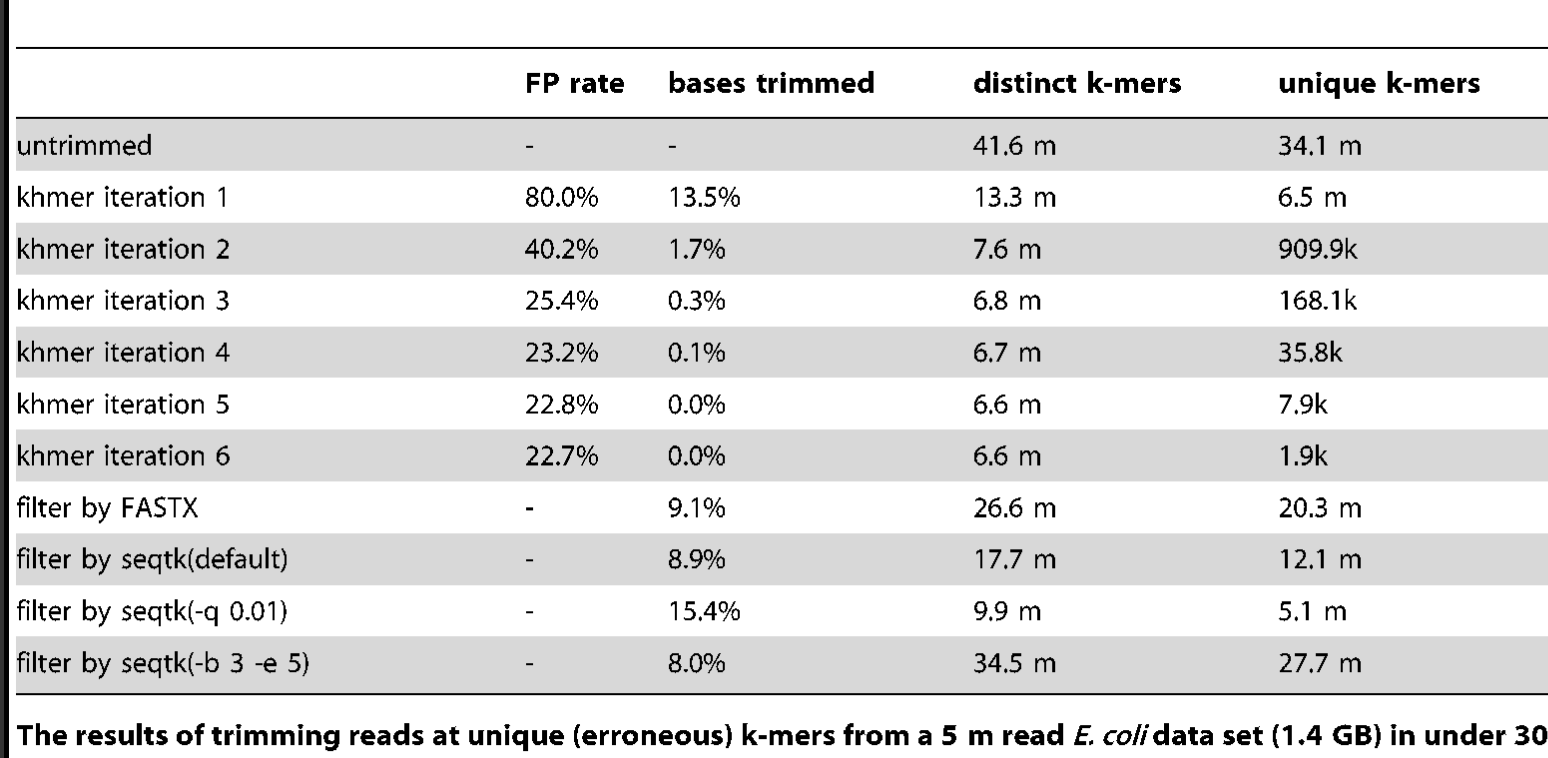
The basic logic is this: if you see low abundance k-mers in a high coverage data set, those k-mers are almost certainly the result of errors. (Caveat: strain variation could also create them.)
In metatranscriptomic data sets we do have the problem that we may have very low and very high coverage data. So we don’t necessarily want to get rid of all low-abundance k-mers, because they may represent truly low abundance (but useful) data.
As part of the khmer project in our lab, we have developed an approach that sorts reads into high abundance and low abundance reads, and only error trims the high abundance reads.
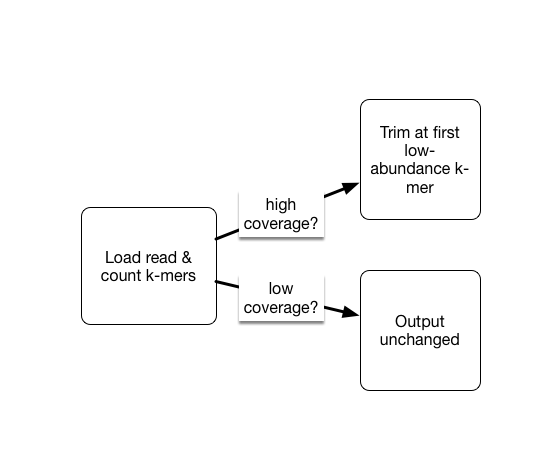
This does mean that many errors may get left in the data set, because we have no way of figuring out if they are errors or simply low coverage, but that’s OK (and you can always trim them off if you really care).
Kmer trimming with Khmer
To properly compare our TARA samples, it would be best to remove these sequence errors. This can also speed up assembly and reduce memory requirements (although many assemblers have built in k-mer trimming mechanisms as well).
Set up workspace and install khmer
We've already installed khmer for you, but here's the command if you need to install it in the future:
conda install khmer
Make sure you have the $PROJECT variable defined:
echo $PROJECT
if you don't see any output, make sure to redefine the $PROJECT variable.
Now, let's create a directory to work in:
cd ${PROJECT}
mkdir -p khmer_trim
cd khmer_trim
Let's choose a sample to start with: TARA_135_SRF_5-20
And link in the qc trimmed files.
ln -s ${PROJECT}/trim/TARA_135_SRF_5-20_*qc.fq.gz ./
Run Khmer
To run error trimming, use the khmer script trim-low-abund.py:
for filename in *_1.qc.fq.gz
do
#Use the program basename to remove _1.qc.fq.gz to generate the base
base=$(basename $filename _1.qc.fq.gz)
echo $base
interleave-reads.py ${base}_1.qc.fq.gz ${base}_2.qc.fq.gz | \
trim-low-abund.py - -V -Z 10 -C 3 -o - --gzip -M 8e9 | \
extract-paired-reads.py --gzip -p ${base}.khmer.pe.fq.gz -s ${base}.khmer.se.fq.gz
done
Assess changes in kmer abundance
To see how many k-mers we removed, you can examine the distribution as above,
or use the unique-kmers.py script. Let's compare kmers for one sample.
unique-kmers.py TARA_135_SRF_5-20_rep1_1m_1.qc.fq.gz TARA_135_SRF_5-20_rep1_1m_2.qc.fq.gz
unique-kmers.py TARA_135_SRF_5-20_rep1_1m.khmer.pe.fq.gz
The first two files (the adapter-trimmed inputs) have a total estimated number of unique 32-mers of 26760613; the second (trimmed) file has a total estimated number of unique 32-mers 26659070. So the trimming removed approximately 100,000 k-mers.
These numbers are virtually the same BECAUSE WE ARE USING SMALL SUBSET DATA SETS. For any real data sets, the second number will be MUCH smaller than the first, indicating that many low-abundance k-mers were removed as likely errors.
For more info and some neat visualization of kmer trimming, go here